Abstract
Myelodysplastic syndromes (MDS) are a heterogeneous clonal bone marrow disorders occurring mostly in the elderly. The Revised International Prognostic System (IPSS-R) groups MDS patients according to the risk of transformation into acute myeloid leukemia. For treatment decision-making, patients are usually divided into two groups: lower- and higher-risk with the cutpoint of 3.5 according to the IPSS-R. However, part of lower-risk MDS patients (LR-MDS) progresses rapidly. Recently, we demonstrated that DNA damage response (DDR) activation and the ensuing senescence form an anti-tumor barrier in CD34+ cells of LR-MDS patients, and that this barrier can be breached by RUNX1 mutations (Kaisrlikova M, et al., Leukemia. 2022; 36:1898). As an extension, we analyzed transcriptomes of LR-MDS patients regardless of their driver mutation, with and without rapid progression (within 2 years of diagnosis), to determine molecular mechanisms underlying the progression. Because MDS is associated with aging, and many anti-tumor barrier mechanisms (DDR, checkpoint activation, senescence) lead to a pro-aging phenotype, we questioned whether aging-related gene expression signatures define the likelihood of disease progression in patients with LR-MDS.
We performed RNA-seq on CD34+ cells of 61 LR-MDS diagnostic samples (53 patients without rapid progression (NRP) and 8 patients with rapid progression (RP)) with a median age of 65 years.
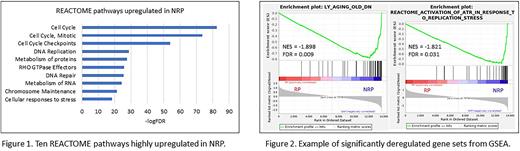
The differential expression analysis showed 1,701 up- and 1,569 downregulated genes in NRP (FDR<0.01). According to the GO Biological Processes (GO BP) and REACTOME databases, the most significantly upregulated pathways were related to macromolecular metabolic processes such as DNA repair, replication, protein metabolism, cell cycle, chromosome organization, including regulation of gene expression, and telomere organization (Fig. 1). The pathways of rRNA expression regulation, splicing, and RNA metabolism were deregulated as well as the pathways related to apoptosis, cellular senescence and cellular response to stress, immune response, and mitochondrial metabolism. Furthermore, we found deregulation of crucial signaling pathways mediated by NOTCH, RHO GTPases, NF-kB, MAPK, b-catenin, and WNT.
Although 1,569 genes were upregulated in RP, only seventeen GO BP terms consisted of regulation of metabolic processes, cytoskeleton organization, phosphorylation, and signal transduction were upregulated.
The NRP results are consistent with the hallmarks of aging (López-Otín C, et al., Cell. 2013; 153:1194). In GSEA, 36/37 and 120/143 gene sets were upregulated in NRP for the datasets consisting of gene sets chosen according to the keyword 'aging' and 'stress', respectively (Fig. 2). Among them, gene sets associated with replication stress, DNA damage, oxidative stress and unfolded protein response were significant (FDR<0.1, ІNESІ>1.7).
Abnormal splicing is associated with cellular aging. Therefore, we conducted a differential alternative splicing analysis using rMATS. We found significant differences in 1,356 alternative splicing events (FDR<0.05, absolute inclusion level difference>10%) between NRP and RP.
Finally, we performed analysis of healthy controls (n=7) and age-matched NRP (NRP-age) (n=13) samples. We observed 70 up- and 209 downregulated genes in NRP-age (FDR<0.05). Only the splicing pathways were upregulated and the immune pathways downregulated. In GSEA, no gene sets were significant (FDR<0.1) for aging and stress datasets.
To conclude, these results show increased signatures of aging in CD34+ cells from NRP, which correspond to an increased level of the intrinsic ability of CD34+ cells to suppress malignant progression through anti-tumor barrier activation. This is consistent with the current understanding that most aging phenotypes are known to be the products of DDR and tumor suppression. Based on our data, we hypothesize that the progression of LR-MDS patients represents a cell-autonomously and perhaps also non-cell-autonomously-mediated failure of appropriate cellular stress response mechanisms. Our data also show similarity at the transcriptome level between age-matched NRP and healthy controls.
Supported by AZV (NV18-03-00227) and (NU21-03-00565), GA CR (N20-19162S), and MH CZ-DRO (UHKT, 00023736).
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal